Introduction
Definition
Diabetic cardiomyopathy (DbCM) represents a cardiovascular disease (CVD) possessing the properties of generation of structural along with functional aberrations people with diabetes mellitus (DM) who do not also have concomitant conditions including hypertension, valve disease, or coronary artery disease in cardiac tissue (CAD) (1). As per the Framingham Heart Study, women along with men with DM possessed a score of 5 and 2, respectively. Heart failure (HF) has a 4 times higher incidence (2). With a range from 19 to 26%, patients with DM had a higher prevalence of HF (3). A case-controlled research discovered that HF was 1.3 times more common than expected greater in patients with DM compared with non-diabetic patients (4). A robust association between type 1 diabetes (T1D) patients and type 2 diabetes (T2D) patients was observed with glycated hemoglobin A1c (HF) and (HbA1c). Every 1% increase in HbA1c, a 30 along with 80%. The incidence of HF was higher in T1D than in T2D, respectively. These stood apart from additional risk factors (5).To start with the phase of DbCM possess the properties of exhaustive cardiac hypertrophy in addition to mild to moderate fibrosis resulting in abnormalities of systolic along with diastolic function of the heart (6).
Environment in blood results
DbCM
In case of experimental along with clinical studies identification of persistent hyperglycemia, insulin resistance (IR), dysfunctional glucose metabolism, and aberrations in insulin signaling, aberrant observations included the intake of heart inflammation, elevated renin-angiotensin system (RAAS) activity, free fatty acids (FFA), and oxidative stress (OS). Furthermore, abnormal mitochondrial functioning acted as one of the main indicators of metabolic changes that triggered a chain reaction of disease-related events. The three most significant mechanisms of the disease’s pathophysiology, namely, the causes of increased cardiomyocyte cell death, left ventricular (LV) hypertrophy, and cardiac fibrosis, are revealed (7).
Biochemical
Mechanistically, numerous molecular modes have been isolated that aid in the pathophysiological alterations in DbCM (Figure 1) [rev in Ref. (8)].

Figure 1. The many processes involved in diabetic cardiomyopathy are illustrated schematically, courtesy of reference no. (8). AGEs, advanced glycation end products.
Additionally, GPR40, 5’-AMP-activated protein kinase (AMPK) signaling, activation of the mitogen-activated protein kinase (MAPK) signaling, O-GlcNAcylation of cardiac proteins, a reduction in insulin, abnormal protein kinase C, and peroxisome proliferator-activated receptor (PPAR) are all involved.
Epigenetic alterations
The etiology of DbCM may be significantly influenced by the dysregulation of miRNAs, circular RNAs (circRNAs), lncRNAs, DNA methylation, and histone posttranslational modifications (such as acetylation, and methylation), according to a recent research (9). After understanding how epigenetic changes contribute to diabetic kidney disease (DKD), diabetic nephropathy, and the association of epigenetic alterations in NAFLD with CVD (10–12), our goal was to evaluate the epigenetic changes in several biological pathways associated with DbCM.
Methods
In this study, we conducted a narrative review utilizing search engines including PubMed, Google Scholar, Web of Science, Embase, and the Cochrane Review Library. We utilized MeSH terms like DbCM, epigenetics, DNA methylation, histone posttranslational modifications, and histone acetylation. CircRNA, miRNAs, lncRNAs, HDAC1- 6, cardiac remodeling, cardiomyocyte apoptosis, cardiac hypertrophy, left ventricular hypertrophy, PI3K signaling pathways, autophagy, pyroptosis, valproic acid, sodium butyrate, and SGLT2 inhibitors from 1950 to the present.
Results
A total of 6,000 articles were discovered, and 132 articles were chosen for this evaluation. They were not meta-analyzed. Figure 2 shows the selection criteria.
Molecular modes along with epigenetic controlling in DbCM
DbCM along with cardiac remodeling
Normal cardiac alteration in the diabetic heart vis-a-vis DbCM
The DbCM often coexists with a number of vascular disorders. Atherosclerosis may be worse in T2DM for a variety of reasons, such as obesity, dyslipidemia, hyperinsulinemia, oxidative stress, low-grade inflammation, anomalies in the metabolism, such as hyperglycemia and a rise in the production of advanced glycation end products (AGEs), and autonomic dysfunction instability, to name a few. stroke, peripheral artery disease, and heart disease are examples of macroangiopathy consequences, whereas retinopathy, neuropathy, and nephropathy are the hallmarks of microangiopathy. Regarding this, it has been discovered that albuminuria and cardiac strain are clearly related in T1DM patients, but SGLT-2 inhibitors have been reported to protect renal function and avoid HF in T2DM patients. Additionally, DCM takes place in association with cardiovascular risk factors including Obesity in general and hypertension make diagnosis and treatment more challenging. The World Health Organization classified obesity as a major risk factor for the development of T2DM in 2014 as one of the stated that more than 600 million individuals were obese globally. Lipid buildup can happen in the obesity and type 2 diabetes (T2DM) are associated with metabolic imbalance and lipotoxicity, which include the main causes of diabetes-related ventricular dysfunction. Additionally, hyperinsulinemia brought on by obesity stimulates the release of VLDL lipoproteins, de novo lipogenesis (DNL), and the expression of lipogenic genes in the liver. These insulin resistance and cardiovascular risk factors distinguish type 2 diabetes (T2DM) from type 1 diabetes (T1DM), whose etiology is primarily driven by genetic factors [rev in Ref. (13)].
Diastolic dysfunction has been identified as an early functional change in the diabetic myocardium. Between 40 and 75% of people with type 1 or type 2 diabetes are thought to have diastolic dysfunction using tissue Doppler imaging and conventional echocardiography. Systolic dysfunction may continue to develop, although typically at a later stage of the illness, which creates a significant challenge for diagnosis when using canonical echocardiography. However, modest changes in systolic function have been identified in 24% of individuals with DM who do not have CAD or LV hypertrophy using strain evaluation and peak systolic velocity calculations. Diastolic failure may not necessarily be the first functional alteration in diabetics since systolic dysfunction can sometimes be present in diabetics with acceptable diastolic function DbCM, according to a recent study that used systolic strain measurement.
Cardiomyocyte hypertrophy along with fibrosis is a significant characteristic of DbCM. Cardiac fibrosis possesses a considerably robust part in the pathophysiology of the disease in case of the human diabetic heart. Robust collagen fibers are deposited in the interstitial area along with perivascular space in the cardiomyocytes with diabetes (14). The majority of the aggravating collagen pathways fibers (type 1 and type II), the wingless-related integration site (WNT), and transforming growth factor beta (TGF-^) signaling pathways are being deposited (15). In addition, the remodeling of matrix metalloproteinases (MMPs) that results in the deregulated breakdown of extracellular matrix (ECM) in diabetic heart aids further (15, 16). TGF-B1 pathway activation and escalated breakdown of ECM take place secondary to RAAS stimulation, leading to accelerated cardiac remodeling, modulated signaling, and hyperglycemia along with IR (17). Reduction in the accessibility of nitric oxide (NO), oxidative stress (OS), increased cardiac collagen deposition, and levels of fibronectin cause interstitial fibrosis when the TGF-1 signaling pathway is activated in conjunction with insulin signaling dysregulation (18). The use of several clinical and animal studies for cardiac fibrosis present in case of diabetes-triggered HF (15, 18) has made it possible to provide amazing proof.
Enhancement of LV hypertrophy as protracted by the existence of greater LV mass along with its correlation with DM has been extensively corroborated (19). The most important foundation for human cardiac hypertrophy is LV thickening (20). Regarding the overall increased LV mass, Myocardial cell death, cardiac fibrosis, and hypertrophy must be considered (21). Besides cardiac hypertrophy, other parameters aiding in DbCM generation are IR, hyperglycemia in the environment, OS that results in the activation of cardiac hypertrophy genes like ^-myosin heavy chain (^-MHC), atrial natriuretic factor (ANP), and brain natriuretic peptide (BNP) (22). Escalated insulin amounts further stimulate cardiac hypertrophy formation. Insulin-like growth factor (IGF-1) stimulates cardiac hypertrophy formation via extracellular signal-regulated kinase (ERK1/2) along with the phosphatidyl inositol 3-kinase (PI3K) signaling pathways (23). The contribution of DM to the development of cardiomyocyte hypertrophy has been further shown in a number of investigations using animal models of DbCM (24).
Epigenetics controlling of cardiac remodeling in DbCM
Effects of hyperglycemia as well as hyperinsulinemia on miRNAs
MicroRNAs represent non-coding RNAs that control cellular gene expression. Abnormal expression of around 30% (alias 300/1000 total RNAs) has been seen in the heart tissue of DM individuals (25). In DbCM, several miRNAs have been identified to control cardiac hypertrophy and fibrosis. Diabetes-induced mice’s cardiac tissue has been reported to dramatically upregulate miRNA-21- like genes (26). By directly influencing the fork head box protein O3 (Fox3), miRNA-212 has been found to regulate the occurrence of cardiac hypertrophy (27). In DbCM, Rac 1 activated kinase (Pak1) and cell division control protein 42 homolog (Cdc42) expression levels. According to Raut et al., miRNA-30c caused an increase. It has been demonstrated that other miRNAs, namely, miRNA- 181a and miRNA-200c, play a substantial role in cardiac remodeling (27–29). miRNA-199a levels were higher in cardiac hypertrophy (26). Recent studies have demonstrated that blocking miRNA-199a, which is a coactivator of the peroxisome proliferator activated receptor (PPAR), can prevent heart hypertrophy by restoring mitochondrial fatty acid oxidation. The diabetic heart exhibited downregulation of miRNA-30a, miRNA-1, and miRNA- 29b (25). The miRNA 144 and miRNA 133 were the two miRNAs with the most relevance in the pathophysiology of diabetes-modified heart failure, out of all the essential orchestrating miRNAs (29). Singh et al. depicted miRNA- 200c (28) to promote cardiac hypertrophy by modifying the expression of dual specific phosphatase 1 (DUSP1) in DbCM (30).
In a diabetic murine model, miRNA-133a was reduced (31). Additionally, it was demonstrated that miRNA-133 levels improved systolic function in addition to reducing fibrosis by reducing collagen levels (32). A biomarker for myocardial fibrosis called miRNA 21 has been identified (33). Numerous studies have shown that in a hyperglycemic environment and in diabetic hearts. In rat cardiac fibroblasts, miRNA 21 levels are increased, which encourages collagen production and fibroblast growth (34). MiRNA 21 also disrupted the JNK and p38 MAPK signalling pathways, which had a direct impact on the activity of the dual specific phosphatase 8 (DUSP8).
Effects of hyperglycemia and hyperinsulinemia on LncRNAs
Long non-coding RNAs are longer than miRNAs. LncRNAs are a different class of non-coding RNAs that have been linked to several disease pathways (35). The lncRNAs with localization in the nucleus work while cytoplasmic lncRNAs often collaborate with miRNAs to control post-transcriptional gene expression. Several lncRNAs, notably DbCM, which supports cardiac fibrosis and hypertrophy, have been linked to the pathogenesis of CVD Due to miRNA-150 and miRNA-93’s ability to act as sponges for anti-hypertrophic miRNAs, myocardial infarction-associated transcript (MIAT) functions as prohypertrophic lncRNAs. Furthermore, MIAT amounts were greater in the myocardium besides the possessed competitive action with miRNA-24 amounts for controlling TGF-B expression, hence cardiac fibrosis (36). Ablating silencing lncRNA Kcnq1ot1 mitigated TGF-B signaling, hence decreasing fibrotic injuries in diabetic mice (37). Dysregulation of both miRNAs along with lncRNAs results in hyperglycemia-associated myocardial injury.
Effects of hyperglycemia and hyperinsulinemia on histone modifications
Furthermore, it has been discovered that cardiac remodeling in DbCM is significantly influenced by histone changes. Non-specific hindering-dependent. In animal models of diabetic heart disease, inhibition of histone deacetylases (HDACs) has been shown to reduce myocardial hypertrophy and fibrosis through increasing the acetylation of glucose transporter 1 and MAPK-modulated phosphorylation (38). Utilization of specific HDAC3 hampering agents like RGF966 further illustrated the enhancement of cardiac function and reverted the DM stimulated cardiac remodeling in diabetic mice. Thus, it was revealed that RGF966 resulted in the reduction of cardiac hypertrophy by modulating the epigenetics of the ERK1/2 pathways modulated via DUSP5 (39). Conversely, Sirt2 is believed to possess advantageous action on DCM. It causes enhancement from cardiac contractile impairment in case of leptin-deficient db/db mice via a histone Sirt2, a deacetylase, directed a route that indicated its potential to function as a therapeutic molecule in DbCM (40).
Part of epigenetics in controlling cell demise modes in DbCM
Modes of cell demise
In the year 2018, the NCCD cell death nomenclature committee demise described various kinds of totally physiological controlled cell death, which is in general labeled as programmed cell death (PCD). Different kinds of PCD have been detailed, including apoptosis (extrinsic along with intrinsic), autophagy-based cell demise, mitochondrial permeability, transit-guided necrosis, ferroptosis, pyroptosis, parthantos, entotic cell demise, NETotic cell demise, lysosomal-based cell demise, and immunogenic cell demise (41). As per this review, we detailed type 1 cell demise (apoptosis), type II cell demise (autophagy-based), and pyroptosis.
Definitions of cell demise modes are discussed as follows:
(i) Apoptosis
Apoptosis represents an event where there is a stoppage of cell growth along with division, instead of spillage of its constituents into the surrounding microenvironment; however, finally, it results in cell demise. Apoptosis is further known as programmed cell death (or “cellular suicide”).
Apoptosis is a foundational physiological event. Its decontrolling has been correlated with different pathologies along with diseases including immune reactions, toxicity of drugs, infections, tumors, and metabolic conditions. Apoptosis possesses the properties of cell shrinking, chromatin condensations, DNA fragmentation, nuclear fragmentation, and blebbing of the cell membrane.
(ii) Autophagy
The word “autophagy” comes from the Greek words “auto” (self) and “phagy” (to eat). Type II cell demise represents the necessary, controlled, conserved catabolic events that modulate recycling along with the breakdown of various cytoplasmic constituents of the eukaryotic cell. Autophagy plays a considerably significant part in cancer and autophagy correlated proteins (ATG). Till now, the double part of autophagy in cancer propagation along with hampering remains debatable. Autophagy possesses a dynamic tumor repressor or tumor-facilitating part in different stages of cancer generation. At the time of early tumor generation, autophagy results in avoidance of the tumor getting started and hampers cancer propagation through survival pathways along with quality regulatory modes. Nevertheless, once the tumor propagates along with forms an end stage, besides exposure to environmental stress, autophagy works in the form of a dynamic recycling system that aids in the survival along with the growth of generated tumors along with facilitates cancer acceleration via metastasis. Therefore, this pointed out that autophagy modulation might prove to be an efficacious interventional approach regarding cancer therapy.
Kinds of autophagy
Dependent on the administration approach, three kinds of autophagy have been detected toward the lysosome, namely, (i) macroautophagy, (ii) microautophagy, and (iii) chaperone-modulated autophagy (CMA) (42). Macroautophagy is the maximum function along with the properties of kinds of autophagy implicated creating a double membrane autophagosome that results in the elimination of injured organelles or undesired cellular constituents by administration to lysosomes for breakdown along with recycling. Different studies have documented that macroautophagy and macroautophagic cell demise represent antitumor reactions. In case of microautophagy, the cargo comprised of (organelles or cytoplasmic constituents) crosstalk directly undergoes fusion with lysosomes for breakdown along with recycling. Microautophagy is more particular in contrast to macroautophagy besides results in the transmission of signals of molecules existent on the surface of injured small organelles like mitochondria and peroxisomes, resulting in particular fusion among lysosomes and these organelles. Based on the organelles whose targeting takes place, the produced autophagic vesicles are labeled by particular names. Mitochondria, peroxisomes, lipids, and RNA are labeled as smitophagy, peroxophagy, lipophagy, and ribophagy, respectively. Intriguingly, CMA reflects the chaperone based selection of cytoplasmic proteins meant for targeting by lysosomes, and their translocation takes place across the membranes of lysosomes for breakdown. A distinctive characteristic of this kind of autophagy is the direct transfer of these proteins without the requirement of selectivity of the proteins broken down besides the generation of extra vesicles. The upregulation of CMA is correlated with cancer cells survival besides proliferation.
(iii) Pyroptosis
Pyroptosis is the inflammation-stimulated cell demise.
DbCM possesses a robust correlation with greater cardiomyocyte cell demise.
Apoptosis along with autophagy represents the considerably significant decontrolled modes implicated in this event (43). Cardiomyocytes, fibroblasts, and endothelial cells have all shown significantly higher rates of apoptosis in the cardiac tissue of individuals with DbCM. Cardiomyocyte death rates were at their highest, and they were followed by fibroblasts and endothelial cells in that order (44). Enhanced cardiomyocyte cell demise causes cell depletion cardiomyopathy and cardiac failure are caused by heart remodelling such cardiac hypertrophy and fibrosis (45).
Heart remodelling, Cardiomyopathy and heart failure are brought on by conditions, such as cardiac fibrosis and hypertrophy (45) are posited to be the main factors responsible for triggering cardiomyocytes cell demise in diabetic hearts (46). Hyperglycemia modulates these effects via local escalated angiotensin II (AngII) (47). More recently, Kobayashi et al. (48) have demonstrated that hyperglycemia might further stimulate cardiomyocytes cell demise via induction of lysosomal membrane permeability along with escalated Heart remodelling, Cardiomyopathy and heart failure are brought on by conditions, such as cardiac fibrosis and hypertrophy (45).
Epigenetics controlling of apoptosis
The expression of numerous miRNAs got dysregulated in hyperglycemia-stimulated cardiomyocytes apoptosis and diabetic heart (26). This includes the expression of miRNA-30c, miRNA-483-3p, and miRNA-181. Among these are miRNA-378, miRNA-34a, miRNA-1, miRNA- 195, and miRNA-144 are some examples of miRNAs. It was shown that H9c2 cardiomyocytes treated with hyperglycemia had elevated miRNA-1, which accelerated apoptosis. They proved that miRNA-1 encourages cardiomyocyte death by suppressing the expression of IGF1. Increased IGF1 miRNA-1 Expression was demonstrated to inhibit the release of cytochrome c induced by glucose and to increase apoptosis by modulating IGF1 (47). Abundant miRNA-34a expression was seen in cardiomyocytes besides controlling a number of proteins, including as the prosurvival protein Sirtuin 1, are expressed (SIRT 1). When cardiomyocytes were given glucose, diabetic hearts and miRNA-34a were found to be upregulated treatment. Fomison Nurse et al. (49) showed that in hyperglycemia- treated cultured cardiomyocytes, the elevation of SIRT 1 was downregulated by miRNA-34a, and miRNA-34a caspase activity that encourages apoptosis was enhanced. The observed decrease in hyperglycemia-stimulated cardiomyocyte apoptosis caused by miRNA-34a interference points to the possibility of a therapeutic (49) component Through the inhibition of its target gene IGF1, miRNA-483-3p was shown by Qiao et al. (50) to be involved in hyperglycemia- stimulated cardiomyocytes’ death. MiRNA-483-cleaved 3p’s expression has been seen in hyperglycemic cardiomyocytes and diabetic mice (28). The facilitation of cardiomyocytes apoptosis by these miRNAs was by uncontrolled p53-p21 axis (28). That upregulation of miRNA-195 stimulates apoptosis in streptozocin (STZ), as well as leptin- deficient mice type 2 diabetic murine hearts by downregulation of SIRT 1 as well as B cell leukemia 2 (Bcl2) (51). Both hearts and cardiomyocytes exhibited altered miRNA- 144 expression in hyperglycemic conditions. In T2D, the upregulation of miRNA-144-3p was discovered (52). Karolina et al. explained how miRNA-144 regulates IRS1 expression in diabetes. In case of hyperglycemia-treated cardiomyocytes, Song et al. (53) revealed that miRNA- 144 targeted C1q/TNF-related protein [CTRP3/c-Jun-N- terminal kinase (JNK)] pathway along with hampering of miRNA-144 ameliorated cardiomyocytes apoptosis. In a different study, Tan et al. (54) revealed that miRNA-144 was decreased in hyperglycemia-treated cardiomyocytes along with diabetic hearts (54). Significant mitochondria functional enhancement occurred subsequent to the cellular expression of Rac 1 of the Rac family of small GTPases, which in turn regulated apoptosis, and miRNA- 144 together reduced myocyte apoptosis through AMPK phosphorylation besides PPAR7/coactivator-1a (PGC-1a) deacetylation (52). Therefore, additional investigation is needed to determine the precise miRNA component. Changes in the phosphatidyl inositol 3-kinase (PI3K)/AKT protein kinase signaling, which lead to the spread of DbCM, stimulated apoptosis, fibrosis, and hypertrophy of cardiomyocytes (55). Myocardial hypertrophy, fibrosis, and apoptosis were inhibited by the overexpression of miRNA- 203 because it inhibited PIK3CA and PI3K/AKT signaling (55). Recent research has suggested that the miRNA- 532 gene is positively correlated with cardiomyocytic apoptosis in diabetic heart disease. It has been shown that miRNA-532 is elevated in T2D, which decreases its main target, the antiapoptotic protein (ARC). In cardiomyocytes treated with hyperglycemia, miRNA-532 overexpression resulted in proapoptotic caspases being activated as well as the opposite being true (56). Another study found that the heart of diabetes patients expressed less antiapoptotic protein. In the diabetic myocardium, downregulation of myocardial Hsp60 as a result of posttranscriptional manipulation by miRNA-1 and miRNA-206 was a strong proapoptotic signal (57).
Apart from miRNAs, different lncRNAs got isolated that were implicated in the modulation of cardiac cell demise in hyperglycemic or diabetic situations (58). Reduction in the expression of lncRNAs H19 in hyperglycemia- treated cardiomyocytes was seen along with enhanced ventricular function in diabetic mice by impeding reduced apoptosis (59). H19 functioned by inhibiting miRNA-675, which modifies the production of the proapoptotic protein VDAC and speeds up cell death (59). Yang et al. (37) illustrated the enhanced expression of one more lncRNA Kcnq1ot1 in the mice’s diabetic hearts. Furthermore, they illustrated that hampering of lncRNA Kcnq1ot1 enhanced cardiac function besides ameliorating pyroptosis (37). Metastasis lung adenocarcinoma transcript1 (MALAT1) represents one more lncRNA that controls hyperglycemia- stimulated cardiomyocytes apoptosis (60). MALAT1 was further demonstrated to result in the downregulation of. By sponging and activating the inflammasome with the NLR family pyrin domain NLRP3 and TGF-^/Smad, one can increase the levels of miRNA-141 or miRNA- 181a-5p signaling (61). Additionally, EZH2 (enhancer of gene) study has demonstrated that MALAT1 influences the overexpression of ATP-binding cassette transporter A1 (ABCA1) and the death of cardiomyocytes homolog2 (EZH2), a histone methyltransferase (62). Along with DbCM, another lncRNA known as HOTAIR has been demonstrated to offer protection against heart cell death under hyperglycemic circumstances. Along with its cardiac overexpression, HOTAIR was shown to be reduced in the hearts of diabetic mice, which slowed the death of the cardiomyocytes in STZ diabetic mice (63). It has been shown that it may control the levels of miRNA-34a by increasing the antiapoptotic target protein SIRT1 and functioning as competitive endogenous RNAs (ceRNAs). In cardiomyocytes treated with hyperglycemia, another lncRNA, MEG3, is increased. MEG3 causes apoptosis by sponging miRNA-145 and raising proapoptotic levels of PDCD4 (64). Later, NKILA (nuclear factor B interacting lncRNA) was found to be significantly greater in individuals with DbCM, in addition to lower hyperglycemia-driven cardiac cell death caused by in vitro silencing (63). In AC 16 cardiomyocytes treated with hyperglycemia, similar quantities of lncRNAs known as LUCAT1 (lung-associated cancer transcript1) were discovered, and aldosterone synthase (CYP11B2) was downregulated to block decreased hyperglycemia-stimulated cardiomyocyte apoptosis (65).
Three crucial epigenetic mechanisms that control gene expression and related physiological processes are DNA, histone methylation, and acetylation processes. A portion of these mechanisms has not yet been thoroughly examined, but future research suggests that they may play a substantial role. In cardiomyocytes treated with hyperglycemia, HDAC1 controlled the inhibition of IGF1R, according to Yu et al. (66). They showed that, in addition to the link between histone4 and IGF1R being diminished, the correlation between histone4 and p53 HDAC1 is enhanced (66). The apoptosis that hyperglycemia induced in cardiomyocytes was also demonstrated to be inhibited by HDAC1 suppression by raising Diabetic GLUT1 acetylation and decreased caspase 3 activity rats. (67). Further evidence that ER stress is that a substantial DbCM regulator comes from the stimulation of apoptosis in cardiac cells (68). Guo et al.’s findings that SIRT1 activation reduced cardiomyocytes from diabetic rats with ER stress and apoptosis were in agreement with the epigenetic modulation of ER stress in DM-stimulated cardiac apoptosis (67). Puthanveetil et al. (69) showed that Foxo1 controlled the effects of hyperglycemia on the production of Indonesian nitric oxide synthase (iNOS) in cardiomyocytes, which increased protein nitrosylation. In hyperglycemic conditions, Foxo1 regulated caspase 3’s nitrosylation, which led to increased cell death (69).
Epigenetics controlling of autophagy
Autophagy represents a physiological event meant to eliminate or recycle injured cell constituents like organelles, proteins along with metabolites from the cell. It is a significant event meant for the sustenance of cell homeostasis. Suppression and escalation Cardiomyocytes exposed to hyperglycemia and diabetic hearts both exhibit signs of autophagy (70, 71). Increased autophagy was seen in the hearts of diabetic mice given fructose (IC3BII; LC2B-1 ratio, pointed that myocardial autophagy was activated in DbCM as revealed by Mellor et al. (72). Nevertheless, Xie et al. (73) found that OVE26 diabetic mice have reduced myocardial AMPK activity and autophagy [724]. Regarding autophagy in the pathophysiology of DbCM, no amiable agreement has been established (72). Dewanjee et al. (71) who recently reviewed autophagy in diabetic hearts demonstrated that autophagy might work in the form of a sword possessing double edges with the earlier activation aiding in the elimination of injured mitochondria, peroxisomes, along with proteins accrual, as well as enhancing antioxidant transcription factor in the same way as nuclear factor 2 linked to erythroid-2 (Nrf2). However, the cell’s autophagy results in autodigestion, an increase in the production of reactive oxygen species (ROS), and heart damage.
Occasional publications pointed that miRNAs might control diabetes-stimulated autophagy. According to Chen et al. (74), individuals with DM had significantly lower levels of circulatory miRNA 30c. Cardiomyocytes and an animal diabetes model showed comparable outcomes. It was miRNA 30c seen to directly control the expression of Beclin. Hence, miRNA 30c downregulation escalated autophagy by escalating proautophagic Beclin1 expression in diabetic hearts. Moreover, miRNA 30c directly controls Beclin1, hence regulating autophagy in DM (74).
Several studies have shown that lncRNAs have a part in DbCM. LncRNAs and Findings by Feng et al. (75) show that DCM-related factor (DCRF) expression in the DbCM was markedly increased in the diabetic mice model. By sponging miRNA-551-b-5p, they showed that DCRF stimulated cardiomyocyte autophagy by raising protocadherin (PCDH17) expression (75). DbCM showed lower levels of lncRNA H19 expression, as shown by Zhou et al. (76). They discovered that the production of lncRNA H19 inhibits DbCM autophagy by regulating the expression of the Di Ras3 (DIRAS3), a GTP-binding protein, which facilitates mTOR phosphorylation.
Epigenetics controlling of pyroptosis
Pyroptosis, also known as inflammation-stimulated cell death, has been shown to accelerate the removal of cardiomyocyte cells in DbCM (77). miRNA 30d encouraged the pyroptosis of cardiomyocytes in hyperglycemic situations by suppressing Foxo3, and ARC, a protein linked with the cytoskeleton that regulates apoptosis, was its downstream effector activity suppressor that resulted in caspase1 activation and the escalation of proinflammatory molecule. Research by Jeyabal et al. (78) showed that miRNA 9 may have a role in the apoptosis that hyperglycemia stimulates in cardiomyocytes. They demonstrated reduced miRNA 9 expression in human diabetic hearts and in vitro hyperglycemia-treated cardiomyocytes. It has been established that the target protein of miRNA 9 is the inflammatory protein 1 that resembles ELAV (ELAVL1). Jeybal et al. (78) demonstrated that downregulation of ELAVL1 expression and overexpression of miRNA 9 alleviated hyperglycemia-stimulated cardiomyocyte pyroptosis, which indicates the diabetic heart’s antiapoptotic function (78).
Epigenetics controlling of mitochondrial impairment in DbCM
(i) Role of miRNA
Mitochondria possess a key part in the maintenance of metabolism and heart function. DbCM production is caused by the elimination of mitochondrial function (79). Adult cardiomyocyte mitochondria generate the majority of intracellular ATP by oxidative phosphorylation. When DM occurs, the mechanism for generating ATP switches from glucose oxidation in the mitochondria to free fatty acid (FFA) oxidation (80). ROS production is accelerated by defective oxidative phosphorylation (81). Apoptosis of cardiomyocytes was also a result of anomalies in Ca2 + flux (82). This improperly managed Ca2 + flow causes the mitochondrial membrane to undergo increased autophagy (83).
In the tissues of the diabetic heart, miRNAs play a significant role in FA metabolism, that is, reduction of miRNA- 133a in diabetes mouse model’s cardiac tissues (32). As per the mode of action, miRNA-133a regulates the CD36 expression by directly controlling testicular protein4 (84). This reasons out the escalation of CD36 expression in the diabetic rat hearts (85). Peroxisome proliferator activated receptor alpha (PPAR a) controls the FFA oxidation in cardiomyocytes (86). Direct controlling of PGCA-a by miRNA-29a takes place (87). Reduction of miRNA-29a amounts was documented in diabetic rat hearts, which further reasoned out the escalated FFA oxidation modulated by PPAR a (88). miRNA-210a amounts were revealed to be 2.5 times greater in human diabetic cardiac failure contrasted to non-diabetic cardiac failure (89). miRNA-210a directly controls acotinase, and complex1 activity is regulated by the iron sulfate complex protein ISCU1/2, which also directs the electron transport chain (ETC) (90). A different team found that the level of miRNA-141 increased in T1D hearts (91). miRNA-141 regulates the soluble carrier family25 members, which in turn regulates the inorganic phosphate shift in the mitochondria 3 (SLC25A3), which in turn impacts how mitochondria make ATP (88). This is similar to how animals with STZ-induced diabetes had increased levels of miRNA-378, which adversely regulates ATP synthase, in their interfibrillar mitochondria (92). All of these investigations indicated that miRNAs play important roles in energy consumption and mitochondrial function in the diabetic heart.
(ii) Role of histone modifiers
Histone modifiers including deacetylases along with acetyltransferases control acetylation at the global level in different physiological conditions of the cell. Thus, the sustenance of homeostasis is attained by them. Adult cardiomyocyte mitochondria generate the majority of intracellular ATP by oxidative phosphorylation. When DM occurs, the mechanism for generating ATP switches from glucose oxidation in the mitochondria to free fatty acid (FFA) oxidation (80). ROS production is accelerated by defective oxidative phosphorylation (81). Apoptosis of cardiomyocytes was also a result of anomalies in Ca2 + flux (82). This improperly managed Ca2 + flow causes the mitochondrial membrane to undergo increased autophagy (83). HDAC hampering in the murine model of DM caused escalated expression of PPAR a that led to the reduction of PPAR-7 that pointed to the part of HDAC amelioration in controlling PPAR fatty acids oxidation in DbCM (93).
Oxidative stress of mitochondria along with its epigenetics controlling
(i) Role of miRNA
The etiology of DbCM involves oxidative stress and the spread of DbCM caused by an increase in IR in cardiomyocytes (Figure 3).

Figure 3. The molecular pathways interactome in the pathogenesis of diabetic cardiomyopathy (DbCM), courtesy reference no. (8). AGEs, advanced glycation end products; ROS, reactive oxygen species.
The ROS are generated at the time of oxygen metabolism in the form of a byproduct. During abnormal situations like insulin resistance (IR) and hyperglycemia, an escalation of nicotinamide adenine nucleotide phosphate (NADPH) oxidases takes place in the mitochondrial respiratory chain that causes shunting of ETC at complex III, which results in the considerable formation of ROS (94). Greater amounts of NADPH oxidases’ action are observed in cardiomyocytes of patients with obesity and cardiac IR (42). This escalated NADPH oxidases result in enhanced ROS generation. Furthermore in DbCM, ROS amounts are enhanced secondary to enhanced xanthine oxidase action besides nitric oxide (NO) uncoupling (95). Mitochondrial impairment results in enhanced ROS accrual. The main factor that determine. In addition to lower oxygen levels, individuals with DbCM also produce hydrogen peroxide (H2O2), hydroxyl radicals, and superoxide molecules (95).
Many miRNAs, including miRNA-1, miRNA-19b, and miRNA 144, have been linked to OS (26, 31). When cardiomyocytes are exposed to a lot of glucose reduction of miRNA-1 amounts occurred along with N-Acetyl cysteine (NAC) treatment that result in cardiac phenotype rescue that corroborated the part of this miRNA in OS-based DbCM (96). The level of miRNA-144 was observed to go into downregulation in hyperglycemic circumstances. By controlling the expression of Nrf2, miRNA-144 directly influences ROS levels (31). The mitochondrial phosphate transporter (Slc25a) was also inhibited by an increase in miRNA-141 in the heart of T1D animals, which increased ROS and lowered mitochondrial ATP synthesis (91). Furthermore, it has been discovered that miRNA-210 regulates mitochondrial metabolism by focusing on the compounds linked to ROS formation (96). As per a different work, reduction of miRNA-373 amounts in DbCM occurred in view of the glucose-stimulated OS-modulated cardiac hypertrophy (97).
(ii) Role of lncRNA
Occasional lncRNAs have been documented to control diabetes-stimulated OS. Downregulation of lncRNA H19 was illustrated in the diabetic rat hearts along with forced expression resulting in the amelioration of OS which in turn subsequently lessened LV impairment (57). Histone DNA deacetylases, one kind of epigenetic modulator, have been further observed to be implicated in OS- modulated pathophysiology of DbCM. Kumar et al. (98) revealed that dyscontrolled SIRT1 and methyltransferases 3b (Dnmt3b) action caused escalated. Diabetes is caused by both CpG demethylation and histone H3 acetylation at the p66Shc (prooxidant adaptor protein) promoter stimulated vascular OS (96). Akin to that, Mortaza et al. (99) explored the modes of reduction of SIRT1 action along with the part of SIRT1 and FOXO3 in ROS-modulated stress (99).
Electrical remodeling along with its epigenetics controlling in the propagation of DbCM
Structural examples like cardiac remodeling (fibrosis and hypertrophy) result in functional aberrations (changed electrical activation) with final electrical remodeling hypertrophy of heart at the time of HF. The properties of electrical remodeling are compensatory or poor adaptability to ongoing ion channel disruptions that may be irreversible or reversible, respectively. Arrhythmias that cause mortality are thought to be caused by this remodeling, which involves the conduction system (100). Although several etiological variables delay the ventricular action potentials, repolarization is a typical type of electrical disturbance that is hypothesized to contribute to the development of CVD remodeling (100). Maximum electrophysiological investigations indicated that the K + currents had decreased possessing a crucial part in electrical remodeling (101). It has been confirmed experimentally that changes in the metabolism of glucose led to the modification of different channels in the ventricle in cardiomyocytes. It would be fascinating to understand more about how the K + channels are changed by DM. Insulin signalling and glucose uptake are both disturbed in cardiomyocytes from DbCM patients with upregulated K + channel activities. It was discovered that insulin therapy is appealing regarding the acquisition of typical transient outer currents in streptozocin (STZ)-induced diabetes in a mouse model (101).
In DbCM (stimulated by type 1 or type 2 DM), prolongation of QT interval is observed (102), escalating the risk of ventricular arrhythmias (103). The expansion of the action potential is primarily governed by the unregulated expression of various ion channel proteins and their characteristics (104).
(i) Role of miRNA
Epigenetic regulators like miRNAs have more recently been found to be involved in the remodeling of the cardiac electrical system (1, 21). Diabetes miRNA-301a controls the expression of the voltage-gated potassium channel Kv4.2 (105). miRNA-29 overexpression caused structural cardiac damage in the diabetic mouse model (106). Increasing the amount of miRNA-141 has an impact on ATP generation by decreasing mitochondrial phosphate transport, as demonstrated by a mouse model of diabetes (91).
(ii) Role of histone modifiers
The function of HDACs in the control of ion channel expression has been emphasized in several publications, although more research is still needed to determine their exact role. In a research, HDAC 5 was used to modulate the sodium-calcium exchanger (NCX)1. This NCX1 is involved in Ca2 + efflux out of the cells, and NK homeobox 5 regulates its expression (NK X2.5). It is connected to the recruitment of HDAC 5 to the promoter of the NCX gene (107). The acetylation of NK X2.5 escalated the crosstalk with HDAC 5, while the deacetylation of NK X2.5 enhanced its affinity toward the p300 complex, which was illustrated by a different study (108). Epigenetic controlling of HDAC 5 impacts Ca2+ flux in the cardiomyocytes (109). Lehmann et al. (110) in this study revealed that the N terminal of HDAC 4 hampers myocyte enhancer factor 2 (MEF2). MEF2 action leads to a reduction in the hexosamine biogenerational pathway and is suppressed by nuclear orphan receptor NR4A expression (110). The porcine model of HF affecting the potassium showed the downregulation of HDAC 2 channels along with QT interval prolongation (111), resulting in the hampering of HDAC 2 along with the influencing action potential. Hampering of HDACs with the utilization of class I hampering agent entinostat might be a feasible therapeutic strategy for HF with the reduction of electrical remodeling besides structural remodeling in HF (112).
Histone deacetylases hampering agent: future therapeutic strategy For DbCM
Histone deacetylases (HDAC) represent the molecules possessing the capacity of pleiotropic functions that are implicated in key homeostasis events like proliferation and cell cycle along with cell demise. HDAC hampering agents (HDACis) in particular block Zn2+ based HDAC enzymes are implicated in histone acetylation. The USFDA recently approved HDACis for cancer therapy in clinical scenario (113). Furthermore, occasional publications pointed that histone acetylation might be an attractive therapeutic modality for the treatment of CVD in the preclinical model (114).
HDACis are classified into 5 categories depending on their structure:
(i) Derivatives of hydroxamic acid, such as panobinostat and trichostatin A
(ii) Short chain fatty (aliphatic) acids, such as sodium butyrate and valproic acid (VPA)
(iii) Cycle-based peptides (like romedepsin)
(iv) Benzamides I (like entinostat)
(v) Sirtuin hampering agents.
These HDACis received the USFDA approval (115). Till date, HDACis utilization has not been done in clinical trials regarding fibrotic diseases; however, the utilization was made for cardiac along with lung fibrosis (116). The biggest actor implicated in this fibrotic situation is the fibroblasts’ transformation into myofibroblasts (117). Studies pointing to HDACis achieving reversion of myofibroblasts activation in animal models have been illustrated. In hypertension, murine model fibrosis was ameliorated by VPA by controlling the acetylation of the corticoid receptor (118). When pressure overload occurred in a mouse model, VPA improved heart remodeling (119). Additionally, VPA reduced cardiac fibrosis via regulating ERK1/2 phosphorylation (120). According to a recent research, VPA reduced the remodeling event, which led to the establishment of atrial fibrillation (121). Similar to how HDACis showed anti-fibrotic effects, MPT0E014 also caused a decrease in Ang II and TGF-receptor expression in a mouse model of cardiomyopathy (122). By increasing the apoptosis and decreasing the phenotype of Ang II myofibroblasts, mocetinostat caused the expression of HDACs to be downregulated in an HF model (123). HDAC 6 silencing or hampering with the utilization of tubacin resulted in the reduction of the expression of TGF-3, hence the reduction of cardiac fibrosis (124). A requirement for greater exhaustive studies is probing into the probability of selective HDACIs for the therapy of DbCM.
(i) Role of sodium-glucose cotransporter 2 (SGLT2) inhibitors
Furthermore, sodium-glucose cotransporter 2 (SGLT2) inhibitors have been observed to be efficacious in the therapy of DbCM (see Figures 4–6 for modes of action apart from epigenetic modes) [rev in Ref. (125)].

Figure 4. Effect of SGLT2 inhibitors on ketone body metabolism, courtesy reference no. (125). By lowering plasma glucose levels and promoting adipose tissue lipolysis and FA synthesis, SGLT2 inhibitor therapy also increases the creation of ketone bodies. In the heart, ketone bodies are metabolized to acetyl- CoA more quickly than glucose or FA. SGLT2 inhibitors also promote the expression of critical genes involved in ketone oxidation, which causes the metabolism to switch using ketone bodies as a preferred substrate. The important metabolites and enzymes are indicated by red arrows, the primary metabolite fluxes are indicated by black arrows, and the synthesis of ATP by the mitochondrial oxidative phosphorylation system is indicated by green arrows. Fatty acids (FA), acetoacetyl CoA, 3-hydroxy-3-methylglutaryl-CoA, hydroxybutyrate, and the tricarboxylic acid cycle are examples of coenzymes. 3-hydroxy- 3-methylglutaryl-coenzyme A lyase is known as HMGCL. Mitochondrial-hydroxybutyrate dehydrogenase, also known as BDH1; adenosine triphosphate or ATP; acetyl-CoA acetyltransferase or ACAT1; acylcarnitine (C2-carnitine); hydroxybutyrylcarnitine (C4-OH carnitine); and succinyl-CoA:3-oxoacid-CoA transferase (CPT1) are examples of carnitine derivatives.

Figure 5. Red arrows point to essential metabolites and enzymes, black arrows point to main metabolite fluxes, and green arrows point to the mitochondrial oxidative phosphorylation system, which produces ATP. Examples of coenzymes include fatty acids (FA), acetoacetyl CoA, 3-hydroxy-3-methylglutaryl-CoA, hydroxybutyrate, the tricarboxylic acid cycle, and 3-hydroxy-3- methylglutaryl-coenzyme. The name HMGCL denotes a lyase. Acylcarnitine (C2-carnitine), hydroxybutyrylcarnitine (C4-OH carnitine), and succinyl-CoA:3-oxoacid-CoA transferase (CPT1) are a few examples of carnitine derivatives. Mitochondrial hydroxybutyrate dehydrogenase, also known as BDH1, acetyl CoA acetyltransferase or ATP. NCLX’s effectiveness, however, is inferior to the MCU’s absorption of Ca2 +. The Ca2 + absorption by SERCA and the leaking from the SR RyR receptors both have an impact on [Ca2 + ]c level. TCA cycle dehydrogenases are controlled by [Ca2 + ]m concentration, which leads to more ATP and less ROS being produced. The major transporters are indicated by red arrows, the fluxes of the ions are indicated by black arrows, and the synthesis of ATP by the mitochondrial oxidative phosphorylation system is indicated by green arrows. Sarcolemmal Na + /H + -exchanger NHE, voltage-dependent L-type calcium channel LTCC, mitochondrial Ca2 + uniporter MCU, sarcolemmal Na + /Ca2 + exchanger NCX, Na + /K + -ATPase NKA, ryanodine receptor RyR, and sarcoplasmic reticulum Ca2 + -ATPase SERCA are some of the other proteins.

Figure 6. Courtesy reference no (125). Beneficial effects of SGLT2 inhibitors on mitochondrial function and the cardiovascular system and the role of diabetes in the development of cardiovascular problems (magenta) (blue).
As mentioned earlier in diabetic nephropathy, SGLT2 inhibitors might have part of epigenetic changes in these modes which need further evaluation (126). SGLT2 inhibitors escalate plasma in addition to tissue amounts of ketone-3 hydroxy butyric acid that stimulated β hydroxy butylation of H3 at Lys 9 of the adiponectin gene in adipocytes, which identifies that a novel histone posttranslational alteration important to the therapeutic activity of SGLT2 inhibitors (52) is independent of their methylation or acetylation. Even if there is limited understanding of DNA methylation, histone epigenetics, or both, there is some knowledge about miRNAs. In an open-label trial, 40 diabetic patients were compared with thiazides or dapaglifiozin for their treatment, and miR30e-5p levels were measured was upregulated whereas miR199 a -3p downregulated in dapaglifiozin treatment patients [135, rev in ref (12)]. This study points out that SGLT2 inhibitors might manipulate epigenetic controllers. Moreover, of the different biomarkers Kumric et al. (127) evaluated, lncRNAs, soluble form of suppression of tumorigenicity 2 along with galectin 3 apparently were of maximum advantages regarding DbCM pickup. Combination of comparatively economical along with precise speckle tracking echocardiography with certain emphasized biomarkers would be an attractive screening approach for the newly diagnosed T2DM. The idea of screening examination would be to guide the influenced patients to more particular corroborating test with the present guidelines which highlight the significance (127) (see Figures 7, 8).

Figure 7. Courtesy reference no. (127). Molecular targets of the diabetic cardiomyopathy biomarkers in cardiomyoctes. MAPK, mitogen-activated protein kinase; PI3K, phosphatidylinositol 3- kinase-protein kinase B; IGFBP7, insulin-like growth factor binding protein 7; GLUT4, glucose transporter type 4.

Figure 8. Courtesy reference no. (127). Molecular target of the soluble form of suppression of tumorigenicity 2. sST2, soluble form of suppression of tumorigenicity 2; IL-33, interleukin-33; ST2L, Suppression of tumorigenicity 2 ligand; TH2, T helper lymphocyte type 2.
Conclusions and future directions
Diabetic cardiomyopathy represents a pleiotropic metabolic disease with complicated etiopathogenesis besides accumulated interactions among genetic and epigenetic factors. In the diabetic environment, there are various cardiomyopathy-inducing factors like ROS-modulated OS hyperglycemic situations, cytokines- modulated inflammation, cell demise (apoptosis, pyroptosis, and autophagy), and epigenetic controlling of dyscontrolled molecular pathways that are stimulated by these mediators. The variations in epigenetic modifications including lncRNAs (miRNA along with lncRNAs), histone modification (methylation and acetylation), and DNA promoter methylation regulate the production of important molecules via many processes that modulate DbCM. In summary, earlier research has shown that genetic, environmental, and epigenetic interactions strongly influence the pathogenesis of DbCM through changes in the cellular signaling system (Figure 9).
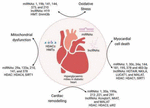
Figure 9. Courtesy reference no. (8). The interaction between several DbCM systems and epigenetic modulators. Long non-coding RNAs, histone deacetylases, histone methyltransferases, microRNAs, lncRNAs, and DNA methyltransferases are few examples.
The past decade has illustrated that miRNA along with lncRNAs are significant controllers of the main molecular pathways like cell demise, mitochondrial impairment, and electrical remodeling. Cardiac fibrosis is a significant event regarding cardiac remodeling event in DbCM. Considerable proof exists regarding the part of epigenetics in diabetic correlated cell demise. Epigenetic controlling modes like histone alterations, DNA methylation, and miRNA along with lncRNAs control cardiac cell demise in the diabetic environment. Akin to that, other modes like mitochondrial impairment and OS are controlled by miRNA along with lncRNAs. Finding these modes has given the provision of generation and formation of innovative therapy approaches for DbCM. miRNAs along with lncRNAs have illustrated translational capacity in the form of diagnostic and prognostic biomarkers, besides therapeutic approaches for DbCM. It was further illustrated that HDACs are significant controllers of the pathophysiology of DbCM. Hampering HDAC with the utilization of hampering agent has demonstrated attractive outcomes regarding cardiac fibrosis. Still the need is present to evaluate controlling modes like chromatin modifications along with circRNAs as ones aiding in the generation of DbCM. Advancements in genomics and molecular biology technologies like transposase accessible chromatin (ATAC) sequences deep sequencing, ChIP sequencing, and high throughput data on DNA methylomes can be made. Provision of a more exhaustive picture of DbCM with this genome-wide data would be feasible. Acquisition of this knowledge will aid in getting insight regarding the part of epigenetic modulators. Furthermore, we already have knowledge regarding the benefits of sodium-glucose cotransporter 2 (SGLT2) hampering agents in the therapy of DbCM. As mentioned earlier in diabetic nephropathy, SGLT2 inhibitors might have part of epigenetic changes in these modes that need further evaluation. SGLT2 inhibitors escalate plasma in addition to tissue amounts of ketone-3 hydroxy butyric acid that stimulated ^-hydroxy butylation of H3 at Lys 9 of the adiponectin gene in adipocytes independent of their methylation or acetylation that isolates a new histone posttranslational modifications significant to the therapeutic action of SGLT2 inhibitors. This study points out that SGLT2 inhibitors might manipulate epigenetic controllers. Moreover, the role of lncRNAs might be of advantage for the early diagnosis of DbCM.
Author contributions
KK and GA reviewed the references. MS finally approved the main draft of the work by KK with mutual agreement.
References
1. Rubler S, Dlugesh J, Yuccoglu Y, Kumral T, Branwood A, Grisham A. New types of cardiomyopathy associated with diabetic glomerulosclerosis. Am J Cardiol. (1972) 30:595–602.
2. Kannel W, Hjortland M, Castelli W. Role of diabetes in congestive heart failure: The Framingham Heart Study. Am J Cardiol. (1974) 34:29–34.
3. Thrainsdottir I, Aspelund T, Thorgeirsson G, Gudnasson V, Hardarson T, Malmberg K, et al. The association between glucose abnormalities and heart failure in the population based Reykjavic study. Diabetes Care. (2005) 28:612–6.
4. Aronow W, Ahn C. Incidence of heart failure in 2737 older persons with and without diabetes mellitus. Chest. (1974) 115:867–8.
5. Lind M, Bounias I, Olsson M, Gudbjornsdottir S, Svennson A, Rosengren A. Glycemic regulation and incidence of heart failure in 20985 patient s with type 1 diabetes mellitus: An observational study. Lancet. (2011) 378:140–6.
6. Westermeir F, Riquelme J, Pavez M, Garrido V, Diaz A, Verdejo H, et al. New molecular insights of insulin in diabetic cardiomyopathy. Front Physiol. (2016) 7:125. doi: 10.3389/fphys.2016.00125
7. Jia G, Hill M, Sowers J. Diabetic cardiomyopathy: A update on mechanisms contributing to the clinical entity. Circ Res. (2018) 122:624–38.
8. Mittal A, Garg R, Bahl A, Khullar M. Molecular mechanisms and epigenetic regulation in diabetic cardiomyopathy. Front Cardiovasc Med. (2021) 8:725532. doi: 10.3389/fcvm.2021.725532
9. Khullar M, Cheema B, Raut S. Emerging evidence of epigenetics modifications in vascular complications of diabetes. Front Endocrinol. (2017) 8:237. doi: 10.3389/fendo.2017.00237
10. Kaur K, Allahbadia G, Singh M. Role of adipocyte impairment in heart failure induction in subjects that are obese along with prediabetes and overt diabetes mellitus: A systematic review. J Cardiol Card Disord. (2021) 2:1–21.
11. Kaur K, Allahbadia G, Singh M. An update on the risk factors correlating NAFLD with cardiovascular disease: Specifically mitochondrial - fatty acids ^-oxidation in liver with therapeutic approaches of avoidance of CVD associated mortality - a systematic review. Open Access J Endocrinol. (2022) 6:1–20. doi: 10.23880/oaje-16000164
12. Kaur K, Allahbadia G, Singh M. Potential role of epigenetic modulation in prevention or therapy for diabetic kidney disease-still a dream or a reality - a systematic review. J Diab Nephro Diab Mgmt. (2021) 1:1.
13. Lorenzo Almoros A, Tunos A, Orejas M, Cortes M, Egido J, Lorenzo O. Diagnostic approaches in diabetic cardiomyopathy. Cardiovasc Diabetol. (2017) 16:28.
14. Shimizu M, Umeda K, Sugihara N, Yoshio H, Ino H, Takeda R, et al. Collagen remodelling in myocardia of patients with diabetes. J Clin Pathol. (1993) 46:32–6.
15. Chiu J, Farhangkhoee H, Xu B, Chen S, George B, Chakrabaru S. PARP mediates structural alterations in diabetic cardiomyopathy. J Mol Cell Cardiol. (2008) 45:385–93.
16. Mizushige K, Yao L, Noma T, Kiyomoto H, Yu Y, Hosomi N, et al. Alterations in left ventricular diastolic filling and accumulation of myocardial at insulin resistant pre-diabetic stage of a type II diabetic rat model. Circulation. (2000) 101:899–907.
18. Jia G, Habibi J, DeMarco V, Martinez-Lemus L, Ma L, Whaley Connell A, et al. Endothelial mineralocorticoids receptor deletion prevents diet induced cardiac diastolic dysfunction in females. Hypertension. (2015) 66:1159–67.
19. Bowden M, Tesch G, Julius T, Rosli S, Love J, Richie R. Earlier onset of diabesity induced adverse remodelling in female compared to male. Obesity. (2015) 23:1166–77.
20. Raut S, Kumar A, Singh G, Nahar U, Sharma V, Mittal A, et al. miR-30c mediates upregulation of Cdc42 and Pak1 in diabetic cardiomyopathy. Cardiovasc Ther. (2015) 33:89–97.
21. Bugger H, Abel ED. Molecular mechanisms of diabetic cardiomyopathy. Diabetologia. (2014) 57:660–71.
22. Dawson A, Morris A, Struthers A. The epidemiology of left ventricular hypertrophy in type2 Diabetes mellitus. Diabetologia. (2005) 48:1971–9.
23. Rosenkranz A, Hood S, Woods R, Dusting G, Richie R. B type natriuretic peptide prevents acute hypertrophic responses in the diabetic rat heart. Diabetes. (2003) 52:2389–95.
24. Sundgren N, Giraud G, Schultz J, Lsarev M, Stork P, Thornburg K. Extracellular signal-regulated kinase and phosphatidyl inositide 3-kinase mediate IGF1 induced proliferation, in fetal sheep cardiomyocytes. Am J Physiol Regul Integr Comp Physiol. (2003) 285:1481–9.
25. Feng B, Chen S, Chiu J, George B, Chakrabaru S. Regulation of cardiomyocyte hypertrophy in diabetes at the transcriptional level. Am J Physiol Endocrinol Metab. (2008) 294:F1119–26.
26. Costantino S, Paneni F, Luscher T, Cosentino F. microRNA profiling unveils hyperglycemic memory in the diabetic heart. Eur Heart J. (2016) 37:572–6.
27. Ucar A, Gupta S, Fiedler J, Ericki E, Kardasinski M, Batkai S, et al. The RNA-212/132 family regulates both cardiac hypertrophy and cardiomyocytes autophagy. Nat Commun. (2012) 3:1078.
28. Raut S, Singh G, Rastogi B, Salika U, Mittal A, Dogra N, et al. miR-30c and miR-181a synergistically p53-p21 in diabetes induced cardiac hypertrophy. Mol Cell Biochem. (2016) 417:191–203.
29. Singh G, Raut S, Khanna S, Kumar A, Sharma S, Prasad R, et al. micro RNA-200c modulates DUSP expression in diabetes induced cardiomyopathy. Mol Cell Biochem. (2017) 424:1–11.
30. Yan H, Wang H, Zhu X, Huang J, Li Y, Zhou R, et al. Adeno- associated virus mediates delivery of miRNA-199a tough decoys attenuates cardiac hypertrophy by targeting PGC-1 alpha. Mol Ther Nucleic Acids. (2021) 23:406–17.
31. Yu M, Liu Y, Zhang B, Shi Y, Cui L, Zhao X. Inhibiting micro RNA-144 abates oxidative stress and reduces apoptosis in hearts of streptozocin induced diabetic mice. Cardiovasc Pathol. (2015) 24:375–81.
32. Chen S, Puthanveeti B, Feng B, Matkovih M, Dorn G, Chakrabaru S. Cardiac miRNA-133a over expression prevents early cardiac fibrosis in diabetes. J Cell Mol Med. (2014) 18:415–21.
33. Kambis T, Shahshahan H, Ker S, Yadav S, Mishra P. Transgenic expression of miRNA-133a in the diabetic akita heart prevents cardiac remodelling and cardiomyopathy. Front Cardiovasc Med. (2019) 6:45. doi: 10.3389/fcvm.2019.00045
34. Reddy S, Hu D, Zhao M, Blay F, Sandeep N, Ong S, et al. miR21 is associated with fibrosis and right ventricular failure. JCI Insight. (2017) 2:e91625.
35. Statello L, Guo C, Chen L, Huarte M. Gene regulation by long noncoding RNAs and its biological function. Nat Rev Mol Cell Biol. (2021) 22:96–118.
36. Qu X, Du Y, Shu Y, Gao M, Sun E, Luo S, et al. MIAT is a profibrotic long noncoding RNAs governing fibrosis in post infarct myocardium. Sci Rep. (2017) 7:42657.
37. Yang F, Qin Y, Lv J, Wang Y, Che H, Chen X, et al. Silencing long noncoding RNAs Kcnq1ot1 alleviates proptosis and fibrosis in diabetic cardiomyopathy. Cell Death Dis. (2018) 9:1000.
38. Chen Y, Du J, Zhao Y, Zhang L, Lv Q, Zhuang S, et al. Histone deacetylases (HDACs) inhibition improves myocardial function and prevents cardiac remodelling in diabetic mice. Cardiovasc Diabetol. (2015) 14:99.
39. Xu Z, Tong Q, Zhang Z, Wang S, Zheng Y, Liu Q, et al. Inhibition of HDAC-3 prevents diabetic cardiomyopathy in OVE26 mice via epigenetic regulation of DUSP ERK1/2 pathways. Clin Sci. (2017) 131:1841–57.
40. Dong F, Ren J. Fidarestat improves cardiomyocyte contractile function in db/db mice through a histone deacetylases Sirt2 dependent mechanism. J Hypertens. (2007) 25:2138–47.
41. Galluzzi L, Vitale I, Aaronson S, Abrams J, Adams D, Agostinis P, et al. Molecular mechanisms of cell death: Recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. (2018) 25:486–454.
42. Jia G, Habibi J, Arour A, Hill M, DeMarco V, Lee L, et al. Enhanced endothelial epithelial cells channels signaling prompts left ventricular diastolic dysfunction in obese female mice. Metabolism. (2018) 78:69–79.
43. Christidi E, Brunham L. Regulated cell death pathways in doxorubicin induced cardiotoxicity. Cell Death Dis. (2021) 12:339.
44. Frustaci A, Kajstura J, Chumenti C, Jaconiuk L, Leri A, Maseri A, et al. Myocardial cell death in human diabetes. Circ Res. (2000) 87:1123–32.
45. Hu X, Bai T, Xu Z, Liu Q, Zheng Y, Cai I. Pathophysiological fundamentals in diabetic cardiomyopathy. Compr Physiol. (2017) 7:693–711.
46. Boudina S, Abel ED. Diabetic cardiomyopathy: Causes and effects. Rev Endocr Metab Disord. (2010) 11:31–9.
47. Fiordaliso F, Li B, Latini R, Sonnebruick E, Anversa P, Leri A, et al. Myocyte death in streptozocin induced diabetes is angiotensin II dependent. Lab Investig. (2000) 80:513–27.
48. Kobayashi S, Zhao F, Kobayashi T, Hagiwara M, Kaminaris A, Li C. Hyperglycemia induced cardiomyocytes cell death is mediated by lysosomal membrane injury and aberrant expression of cathepsin D. Biochem Biophys Res Commun. (2020) 523:239–45.
49. Formison Nurse I, Saw EE, Gandhi S, Munasinghe P, van Hout I, Williams M, et al. Diabetes induces the activation of proageing miRNA-34a in the heart but has differential effects on cardiomyocytes and cardiac progenitor cells. Cell Death Differ. (2018) 25:1330–49.
50. Qiao Y, Zhao Y, Liu Y, Ma N, Wang C, Zou I, et al. miR-483- 3p regulates hyperglycemia induced cardiomyocytes apoptosis in Transgenic mice. Biochem Biophys Res Commun. (2016) 477:541–7.
51. Zheng D, Ma J, Yu Y, Li M, Ni R, Wang G, et al. Silencing of miR- 195reduces diabetic cardiomyopathy in C57BL/6mice. Diabetologia. (2015) 58:1949–58.
52. Karolina D, Arnugam A, Tavintharan S, Wong M, Lim S, Sum C, et al. microRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrates 1 in type2 diabetes mellitus. PLoS One. (2011) 6:e22839. doi: 10.1371/journal.pone.0022839
53. Song Y, Mai H, Lin Y, Wang Y, Wang X, Gu S, et al. miR 144 affects proliferation and apoptosis of high glucose inducedAC16 cardiomyocytes by regulating CTRP3/JNK signaling. Int J Clin Exp Med Pathol. (2020) 13:142–52.
54. Tan L, Huang X, Xu M, Yang L, Hua F. miR 144 protects the heart from hyperglycemia induced injury by regulating mitochondria biogenesis and cardiomyocytes apoptosis. FASEB J. (2020) 34:2173–97.
55. Yang X, Li X, Lin Q, Xu Q. Upregulation of microRNA 203 inhibits myocardial fibrosis and Oxidative stress in mice with diabetic cardiomyopathy through the inhibition of PI3K/Akt signaling pathway via PIK3CA. Gene (2019) 715:143995.
56. Chandrasekra D, Neale P, van Hout I, Rawal S, Coffey S, Jones G, et al. Upregulation of microRNA 532enhances cardiomyocytes apoptosis in the diabetic heart. Apoptosis. (2020) 25:388–99.
57. Shan Z, Lin QX, Deng C, Zhu J, Mai I, Liu J, et al. miR1/miR206 regulates Hsp60 contributing to glucose mediated apoptosis in cardiomyocytes. FEBS Lett. (2010) 584:3592–600.
58. Guo Y, Feng X, Wang D, Kang X, Zhang L, Ren H, et al. Long noncoding RNA: A key regulator in the pathogenesis of diabetic cardiomyopathy. Front Cardiovasc Med. (2021) 8:655598. doi: 10.3389/fcvm.2021.655598
59. Li X, Wang H, Yao B, Xu W, Chen J, Feng Zhou X. LncRNA H19/miR-675 axis regulates cardiomyocytes apoptosis by VDAC1 in diabetic cardiomyopathy. Sci Rep. (2016) 6:36340.
60. Zhang M, Gu H, Xu W, Zhou X. Downregulation of lncRNA MALAT1 reduces cardiomyocytes apoptosis and improves left ventricular function in diabetic rat. Int J Cardiol. (2016) 203:214–6.
61. Che H, Wang Y, Li H, Li Y, Sahil A, Lv I, et al. Melatonin alleviates cardiac fibrosis via inhibiting lncRNA MALAT1/miR-141 mediated NLRP3 inflammasome and TGF-6/Smads signaling in diabetic cardiomyopathy. FASEB J. (2020) 34:5282–98.
62. Wang C, Liu G, Yang H, Guo S, Wang H, Dong Z, et al. MALAT1 mediated recruitment of the histone methyltransferase EZH2 to the microRNA 22 promoter leads to cardiomyocytes apoptosis in diabetic cardiomyopathy. Sci Total Environ. (2021) 766:142191.
63. Gao I, Wang X, Guo S, Xiao I, Liang C, Wang Z, et al. LncRNAs HOTAIR functions as a to competing endogenous RNAs lncRNAs HOTAIR competing endogenous RNAs to upregulate SIRT1 by upregulating miR-34a in diabetic cardiomyopathy. J Cell Physiol. (2019) 234:4944–58.
64. Chen Y, Zhang Z, Zhu D, Zhao W, Li F. Long noncoding RNA MEG3 serves as a ceRNA for miRNA-145 to induce apoptotis of AC16 cardiomyocytes under high glucose conditions. Biosci Rep. (2019) 39:BSR20190444.
65. Li Q, Li P, Su J, Liu S, Yang X, Yang Y, et al. LncRNAs NKILA was upregulated in diabetic cardiomyopathy with early prediction values. Exp Ther Med. (2019) 8:1221–5.
66. Yu X, Geng Y, Liang J, Lin Q, Lin S, Zhang S, et al. High levels of glucose induce apoptosis in cardiomyocytes via epigenetic regulation of the insulin like growth factor receptor. Exp Cell Res. (2010) 316:2903–9.
67. Guo R, Liu W, Liu B, Zhang B, Li W, Xu Y. SIRT1 suppresses cardiomyocytes apoptosis in diabetic cardiomyopathy: An insight into Endoplasmic Reticulum stress response mechanism. Int J Cardiol. (2015) 191:36–45.
68. Ozcan U, Cao Q, Yilmaz F, Lee A, Iwakoshi N, Ozdelen E, et al. Endoplasmic reticulum stress links Obesity, insulin action and type 2 diabetes. Science. (2004) 306:457–61.
69. Puthanveeti F, Zhang D, Wang Y, Wang F, Wan A, Abraham A, et al. Diabetes triggers PARP1 mediated cell death pathway in the heart through participation of FoxO3. J Mol Cell Cardiol. (2012) 53:677–86.
70. Kanamori H, Takemura G, Goto S, Tsujimoto A, Mikaram A, Ogi no A, et al. Autophagic in diabetic cardiomyopathy differ between type1 and type2 Diabetes. Autophagy. (2015) 11:1146–60.
71. Dewanjee S, Vallamkondu I, Kalra R, John A, Reddy P, Kandimalla R. Autophagy in the diabetic heart: A potential pharma- cotherapeutic target in diabetic cardiomyopathy. Ageing Res Rev. (2021) 68:101338.
72. Mellor K, Bell T, Young M, Richie R, Delbridge I. Myocardial autophagy activation and suppressed survival signaling is associated with insulin resistance in fructose fed mice. J Mol Cell. (2011) 50:1035–43.
73. Xie Z, Lau K, Eby B, Lozano P, He C, Penningto B, et al. Improvement of cardiac function by chronic metformin treatment is associated with enhanced cardiac autophagy in diabetic OVE26 mice. Diabetes. (2011) 60:1770–8.
74. Chen C, Yang X, Li H, Yin Z, Fan I, Zhao Y, et al. miR 30c is involved in diabetic cardiomyopathy through regulation of cardiac autophagy via BECN1. Mol Ther Nucleic Acids. (2017) 7:127–39.
75. Feng Y, Xu W, Zhang W, Wang W, Liu C, Zhou X. LncRNA DCRF regulates cardiac autophagy via miR-551-b-5p in diabetic cardiomyopathy. Therapeutics. (2019) 9:4558–66.
76. Zhou C, Jiang R, Liu X, Shao M. LncRNA H19 initiates autophagy by epigenetically silencing of DIRAS3 in diabetic cardiomyopathy. Oncotarget. (2016) 8:1429–37.
77. Li X, Du N, Zhang Q, Li J, Chen X, Liu X, et al. miRNA 30d regulates cardiomyocytes pyroptosis by directly targeting Foxo3 in diabetic cardiomyopathy. Cell Death Dis. (2014) 5:e1479.
78. Jeyabal F, Thandavarayan R, Joladarahi D, Suresh Babu S, Krishnamurthy S, Bhimaraj A, et al. miRNA-9 Inhibits hyperglycemia induced pyroptosis in human ventricular cardiomyocytes by targeting ELAVL1. Biochem Biophys Res Commun. (2016) 471:423–79.
79. Kim J, Wei Y, Sowers J. Role of mitochondria dysfunction in insulin resistance. Circ Res. (2008) 102:401–14.
80. Jia G, DeMarco V, Sowers J. Insulin resistance and hyper- insulinemia in diabetic cardiomyopathy. Nat Rev Endocrinol. (2016) 12:144–53.
81. Anderson E, Kypson E, Rodriques F, Anderson C, Lehr F, Necfer P. Substrate specific derangements in mitochondrial metabolism and redox balance in the atrium of type 2 diabetes. J Am Coll Cardiol. (2009) 54:1891–8.
82. Anderson E, Rodriques F, Anderson C, Thayne R, Chitwood W, Kirpano A. Increased propensity of cell death in diabetic human heart is mediated mitochondrial dependent pathways. Am J Physiol Heart Clin Physiol. (2011) 300:11118–21.
83. Shaikh S, Troncoso R, Criollo A, Bravo-Sagua R, Garcia I, Morselli E, et al. Regulation of cardiomyocytes autophagy by calcium. Am J Physiol Endocrinol Metab. (2016) 300:E587–96.
84. Feng X, Huang I, Liu Z. miRNA-133a attenuates lipid accumulation via TR4-CD36 pathway. Biochimie. (2016) 127:79–85.
85. Pelsers M, Lutgerink T, vanNieuweniunve F, Tandon N, van der Vusse G, Arends J, et al. A sensitive immunoassay for the rat fatty acid translocase (CD36) using phage antibodies selected transfectants: Abundant presence of fatty acid translocase (CD36) in cardiac and red skeletal muscle and upregulation in Diabetes. Biochim J. (1999) 337:407–14.
86. Liang F, Wang F, Garder D. Peroxisome proliferator activated receptor alpha (PPAR a) agonists inhibit hypertrophy of neonatal rat cardiac myocytes. Endocrinology. (2003) 144:4187–94.
87. Liang J, Liu C, Qiao A, Cui Y, Zhang H, Cui A, et al. miRNA- 29a decreases fasting blood glucose levels by negatively regulating hepatic gluconeogenesis. J Hepatol. (2013) 58:535–42.
88. Diao X, Shen E, Wang X, Hu B. Differentially expressed microRNAs and their target genes in the heart of streptozocin induced diabetic mice. Mol Med Rep. (2011) 4:633–40.
89. Greco S, Fasanaro P, Castelvecchio S, Alessandra Y, Arcelli D, DiDonato M, et al. microRNA dysregulation in diabetic ischemic heart failure patients. Diabetes. (2012) 61:1633–41.
90. Chen S, Zhang Y, Hemann C, Mahoney C, Zweir J, Loscalzo J. microRNA 210 control mitochondrial metabolism during hypoxia by repressing the iron sulfur cluster assembly proteins ISCU1/2. Cell Metab. (2009) 10:273–84.
91. Baseler W, Thapa D, Jagannathan R, Dabkoswski E, Croston T, Hollander J. microRNA miR 141 as a regulator of the mitochondrial phosphate carrier (Slc25a3) in the type1 diabetic heart. Am J Physiol Cell Physiol. (2012) 303:1244–51.
92. Jagannathan R, Thapa D, Nichols C, Shepherd D, Stricker J, Croston T, et al. Translational regulation of the mitochondrial genome following redistribution of mitochondria microRNA in the diabetic heart. Circ Cardiovasc Genet. (2015) 8:785–802.
93. Lee T, Kao Y, Tsai W, Chung C, Chen Y, Chen Y. HDAC inhibition modulates cardiac PPARs and fatty acids metabolism in diabetic cardiomyopathy. PPAR Res. (2016) 2016:5938740.
94. Teshima Y, Takashi N, Nishio S, Saito S, Kondo H, Fukui A, et al. Production of reactive oxygen species in the diabetic heart: Role of mitochondria and NADPH oxidases. Circ J. (2014) 78:300–6.
96. Yildrim S, Akman D, Catalucci D, Turan B. Relationship between downregulation of miRNAs and increase of oxidative stress in the development of diabetic cardiac dysfunction: Junction as a target protein of miR 1. Cell Biochem Biophys. (2013) 67:1397–408.
97. Magenta A, Green S, Gaetano C, Martelli F. Oxidative stress and miRNAs in vascular disease. Int J Mol Sci. (2013) 14:17319–46.
98. Shen E, Diao X, Wang X, Chen R, Hu B. miRNAs involved during the mitogen activated protein kinase cascade pathway in glucose induced cardiac hypertrophy. Am J Pathol. (2011) 179:6397–6350.
99. Kumar S, Kim Y, Vikram A, Naqvi A, Li Q, Kassan M, et al. Sirtuin 1 regulated lysine acetylation p66Shc governs diabetes induced vascular oxidative stress and endothelial dysfunction. Proc Natl Acad Sci USA. (2017) 114:1714–9.
100. Mortaza R, Chen S, Feng B, Sen S, Chakrabarti S. High glucose induced alterations in SIRT1 in endothelial cells causes rapid aging in a p300 and FOXO regulated pathway. PLoS One. (2013) 8:e54514. doi: 10.1371/journal.pone.0054514
101. Tomaselli G, Marban E. Electrophysiological remodelling in hypertrophy and heart failure. Cardiovasc Res. (1999) 42:270–83.
102. Xu L, Patel K, Rozanski G. Metabolic basis of decreased transient outward K+ currents in ventricular myocytes fromdiabetic rats. Am J Physiol Heart Circ Physiol. (1996) 271(5 Pt. 2):H2190–H2196.
103. Ninkovic V, Ninkovic S, Miloradovic V, Stanojevic D, Babic M, Giga V, et al. Prevalence and risk factors for prolonged QT interval and QT dispersion in patients with type 2 diabetes. Acta Diabetol. (2016) 53:737–44.
104. Hegyi B, Bers D, Bpssuyt J. CaMKII signaling in heart diseases: Emerging role in diabetic cardiomyopathy. J Mol Cell Cardiol. (2019) 127:246–59.
105. Torres Jacome J, Gallego M, Rodriques-Robledo J, Sanchez Cha- Pula J, Casis O. Improvement in the metabolic stress reverses cardiac potassium channels synthesis in experimental diabetes. Acta Physiol. (2013) 207:447–59.
106. Panguluri S, Tur J, Chapalmadugu K, Katnic C, Cuevas J. Tip- paraju SM. miRNA-301a mediated regulation of Kv4.2 in diabetes: Identification of key modulators. PLoS One. (2013) 8:e60545. doi: 10.1371/journal.pone.0060545
107. Arnold N, Koppula P, Gul R, Luck C, Pulakat L. Regulation of cardiac expression of the diabetes marker microRNA miR 29. PLoS One. (2014) 9:e103284. doi: 10.1371/journal.pone.0103284
108. Harris L, Wang S, Mani S, Kasiganesan H, Chou C, Menick D. Evidence for a non canonical role of HDAC 5 in regulation of the Ncx1 and Bnp genes. Nucleic Acids Res. (2016) 44:3610–7.
109. Chandrasekaran S, Peterson R, Mani S, Addy B, Buccholz A, Xu L, et al. Histone deacetylase facilitates Sodium/calcium channels exchanger upregulation in adult cardiomyocytes. FASEB J. (2009) 23:3851–64.
110. Lehmann L, Jebessa Z, Kreusser M, Hursch A, He T, Kronlage M, et al. A proteolytic fragment 4 of histone deacetylase 4 protects the heart from failure by regulating the hexosamine biosynthetic pathway. Nat Med. (2018) 24:62–72.
111. Syren P, Rahim A, Schweizer P, Bruehl C, Katus H, Frey N, et al. Histone deacetylase 2 dependent ventricular electrical remodelling in a porcine model of early heart failure. Life Sci. (2021) 281:119769.
112. Freundt J, Frommeyer G, Spieker T, Wotzel E, Grotthoff J, Stypmann J, et al. Histone deacetylase inhibition by entinostat for prevention of electrical and structural remodelling in heart failure. BMC Pharmacol Toxicol. (2019) 20:16. doi: 10.1186/s40360-019-0294-x
113. Qin H, Li H, Liu F. Selective histone deacetylase small molecule inhibitors: Recent progress and perspectives. Expert Opin Ther Pat. (2017) 27:621–30.
114. Kim H, Rowe M, Ren M, Hong J, Chen P, Chang D. Histone deacetylase inhibitors exhibit antiinflammatory and neuroprotec- tive effects in a rat permanent ischemic model of stroke: Multiple mechanisms of action. J Pharmacol Exp Ther. (2007) 321:892–901.
115. Eckschlager T, Plch J, Stiborova M, Hrabeta J. Histone deacetylase inhibitors as anticancer drugs. Int J Mol Sci. (2017) 18:E1414.
116. Li M, Zheng Y, Yuan H, Liu Y, Wen X. Effects of dynamic changes in histone acetylation and deacetylase activity on pulmonary fibrosis. Int Immunopharmacol. (2017) 52:272–80.
117. Ku K, Liu X, Hagood J. Myofibroblasts differentiation and survival in fibrotic disease. Expert Rev Mol. (2011) 13:e27.
118. Kang S, Senk Y, Song M, Lee H, Kurtz T, Kim I. Histone deacetylase inhibition attenuates cardiac hypertrophy and fibrosis through acetylation of mineralocorticoid receptor in spontaneously hypertensive rats. Mol Pharmacol. (2015) 87:782–91.
119. Liu Y, Li S, Zhang Z, Jiang H, Lv Z, Tan X, et al. Effects of valproic acid on sympathetic activity and left ventriculo myocardial remodelling in rats during pressure overload. Turk J Med Sci. (2017) 47:1651–60.
120. Zhang Y, Gao F, Tang Y, Xiao J, Li C, Ouyang Y, et al. Valproic acid regulates Ang II induced pericyte myofibroblasts transdifferentiation via MAPK/ERK pathway. Am J Transl Res. (2018) 10:1976–80.
121. Schuliz R, Schuliz J, Hamer S, Himmler K, Puteanu F, Skedl M, et al. Histone deacetylase Inhibitor (HDACIs) valproic acid attenuates atrial remodelling and delays onset of atrial fibrillation in mice. Circ Arrythm Electrophysiol. (2019) 12:e007071.
122. Kao Y, Liou J, Chung C, Lien G, Kuo C, Chen S, et al. Histone deacetylase inhibition improves cardiac function with direct antifibrotic activity in heart failure. Int J Cardiol. (2013) 168:4178–83.
123. Nural Guvener H, Zakharova L, Nimlos J, Popovic S, Mastroent D, Gaballa M. HDAC class1 inhibitor, mocetinostat reverses cardiac fibrosis in heart failure and diminishes CD90+ cardiac myofibroblasts activation. Fibrogenesis Tissue Repair. (2014) 7:10.
124. Tao H, Yang J, Hu W, Shi K, Li J. HDAC 6 promotes cardiac fibrosis progression, through suppressing RASSF1A expression. Cardiology. (2016) 133:18–26.
125. Dabravolski S, Zhuravlev A, Kartuesov A, Borisov E, Sukhorukov V, Orekhov A. Mitochondria mediated cardiovascular sodium-glucose cotransporter 2 inhibitors. Int J Mol Sci. (2022) 23:5371.
126. Solini E, Seghieri M, Giannini L, Biancalana E, Parolini F, Rossi C, et al. The effects of dapagliflozin on systemic and renal vascular function display an epigenetic signature. J Clin Endocrinol Metab. (2019) 104:4253–63.
127. Kumric M, Kurir T, Borovac J, Bozik J. Role of novel biomarkers in diabetic cardiomyopathy. World J Diabetes. (2021) 12:685–705.
128. Liu S, Li W, Xu M, Huang H, Wang J, Chen X. Micro RNA 21 targets dual specific phosphatase-8 to promote collagen synthesis in high glucose treated primary cardiac fibroblasts. Can J Cardiol. (2014) 30:1689–99.
129. Zhu X, Yuan Y, Rao S, Wang P. LncRNA MIAT enhances cardiac hypertrophy partly by sponging through miRNA-150. Eur Rev Med Pharmacol Sci. (2016) 20:3653–60.
130. Li Y, Wang J, Sun L, Zhu S. LncRNA myocardial infarction associated transcript (MIAT) contributed to cardiac hypertrophy by regulating TLR4 via miRNA 93. Eur J Pharmacol. (2018) 818:508–17.
131. Yu X, Song Y, Geng Y, Lin Q, Shan Z, Lin S, et al. Glucose induces apoptosis of cardiomyocytes via miRNA-1 and IGF1. Biochem Biophys Res Commun. (2008) 376:548–52.
132. Yin Y, Yang Z, Li X, Zhou L, Zhang Y, Yang B. Knockdown of long noncoding RNA LUCAT1 reverses high glucose induced cardiomyocytes injury via targeting CYP11B2. EurRev Med Pharmacol Sci. (2019) 23:8560–6.
133. Zeng C, Wang R, Tan H. Role of Pyroptosis in cardiovascular diseases and its therapeutic implications. Int J Biol Sci. (2019) 15:1345–57.
135. Wu Y, Leng Y, Meng Q, Xue R, Zhao B, Zhan L, et al. Suppression of excessive histone deacetylases activity in diabetic hearts attenuates myocardial ischemia/reperfusion injury via mitochondrial apoptosis pathway. J Diabetes Res. (2017) 2017:8208065.
